PineAPPL
Tutorial for PineAPPL’s CLI
Welcome to PineAPPL’s CLI tutorial! Here we’ll explain the basics of PineAPPL’s
command-line interface (CLI): that’s the program pineappl that you can you
use inside your shell to convolve grids with PDFs and to perform other
operations. This tutorial will also introduce and explain the terminology
needed to understand the C, Fortran, Python and Rust API.
This tutorial assumes that you understand the basics of interpolation grids. If you’d like to refresh your memory read the short overview.
Prerequisites
To follow the tutorial, you’ll first need PineAPPL’s CLI; if you haven’t installed it yet follow its installation instructions. Next, you’ll need a fresh directory. For instance, run
cd $(mktemp -d)
to create a temporary directory. Finally, you’ll need a grid,
wget 'https://data.nnpdf.science/pineappl/test-data/LHCB_WP_7TEV.pineappl.lz4'
which you’ll use with the CLI.
pineappl convolve: Performing convolutions
Now that you’ve got a grid, you can perform a convolution with a PDF set:
pineappl convolve LHCB_WP_7TEV.pineappl.lz4 CT18NNLO
We chose to use the default CT18 PDF set for this tutorial, because it’s the shortest to type. If you get an error that reads
error: Invalid value for '<PDFSETS>...': The PDF set `CT18NNLO` was not found
install the PDF set with LHAPDF, or use a different PDF set—the numbers won’t matter for the sake of the tutorial. If the command was successful, you should see the following output:
b etal dsig/detal
[] [pb]
-+----+----+-----------
0 2 2.25 7.7526895e2
1 2.25 2.5 7.1092145e2
2 2.5 2.75 6.1876958e2
3 2.75 3 5.0017809e2
4 3 3.25 3.7228440e2
5 3.25 3.5 2.5236943e2
6 3.5 4 1.1857770e2
7 4 4.5 2.7740964e1
Let’s have a closer look at what the output shows:
- the
bcolumn shows there are 8 bins, indexed0through7, - the next two columns labelled
etalshows the observable used to define the bins and its left and right bin limits, - the last column
dsig/detalshows the differential cross section (dsig) for the bin limits given in the previous two columns.
In the case where the grid requires different types of convolution functions,
the LHAPDF names need to be appended with +p, +f, or +pf (equivalently
+fp) which respectively correspond to a polarized PDF, a fragmentation function, or
a polarized fragmentation function.
As an example, let’s download a grid that contains one polarized and one unpolarized proton:
wget 'https://data.nnpdf.science/pineappl/test-data/STAR_WMWP_510GEV_WM-AL-POL.pineappl.lz4'
Then, to perform the convolution, run the following command:
pineappl convolve STAR_WMWP_510GEV_WM-AL-POL.pineappl.lz4 NNPDFpol11_100+p,CT18NNLO
where we specified the PDF set NNPDFpol10_100 to be polarized. Note that
the order in which the PDF sets are passed do not matter; pineappl will
automatically identify which object should be convolved with a given PDF set.
If successful, the command above should print the following result (again the numbers don’t really matter):
b eta dsig/deta
[] [pb]
-+------------------+------------------+-----------
0 -1.5 -1 7.6327588e1
1 -1 -0.5 5.6712977e2
2 -0.5 0 1.8279428e3
3 0 0.4999999999999999 3.5666164e3
4 0.4999999999999999 1 3.9650498e3
5 1 1.5 1.2597799e3
By default, pineappl uses the first PDF set to compute the value of
$\alpha_s(Q^2)$. This can be changed by passing the position of the PDF set as
an index (starting from 0) to the --use-alphas-from flag.
pineappl -h: Getting help
One of the most difficult aspects of learning a new program is remembering how
to achieve certain tasks and what to type. This should be easy with pineappl;
just remember that you can type
pineappl
which is a shortcut for pineappl -h. This will give you a list of program
options and subcommands, under which all operations in pineappl are
grouped, and a corresponding description. You’ll be familiar with the concept
of subcommand if you’re using git: add, commit and push are well-known
subcommands of it.
To get more help on a specific subcommand, for instance convolve, which we’ve
used already, run
pineappl convolve -h
Depending on the version of PineAPPL this will show output similar to the following:
Convolutes a PineAPPL grid with a PDF set
Usage: pineappl convolve [OPTIONS] <INPUT> <PDFSETS>...
Arguments:
<INPUT> Path of the input grid
<PDFSETS>... LHAPDF id(s) or name of the PDF set(s)
Options:
-b, --bins <BINS> Selects a subset of bins
-i, --integrated Show integrated numbers (without bin widths) instead of differential ones
-o, --orders <ORDERS> Select orders manually
--digits-abs <ABS> Set the number of fractional digits shown for absolute numbers [default: 7]
--digits-rel <REL> Set the number of fractional digits shown for relative numbers [default: 2]
-h, --help Print help
This explains that pineappl convolve needs at least two arguments, the first
being the grid file, denoted as <INPUT> and a second argument <PDFSETS>,
which determines the PDF set. Note that this argument has three dots, ...,
meaning that you’re allowed to pass multiple PDF sets, in which case pineappl
will perform the convolution with each PDF set, such that you can compare them
with each other. This subcommand also accepts a few optional parameters,
denoted with [OPTIONS].
pineappl read: What does this grid contain?
If you’re experienced enough in high-energy physics, you’ve already inferred
from the file name of the grid and the observable name etal what the
numbers will most likely show. However, how can you be certain? Specifically,
if you didn’t generate the grid yourself you’ll probably want to know the
answers to the following questions:
- For which process is the prediction for?
- Which observable is shown?
- If there’s a corresponding experimental paper, where is it?
- Where are the measured values corresponding to the shown predictions?
- Which program/generator was used to produce this grid? What version?
- Which runcards were used, which parameter values?
The subcommand that’ll answer all questions is read. It gives you access to
what we call metadata (the data describing the interpolation grids):
pineappl read --show LHCB_WP_7TEV.pineappl.lz4
This will print out a very long list of (alphabetically sorted) key–value pairs, from which you can read off the answers to the questions above. Let’s go through them one by one:
- The value for the key
descriptiongives a short description of the process. In this case this isLHCb differential W-boson production cross section at 7 TeV. - The keys
x1_labelcontains the name of the observable, andy_labelthe name of the corresponding (differential) cross section. These strings are being used byconvolveand other subcommands that perform convolutions to label the columns with the corresponding numbers. If grids contain two- or even higher-dimensional distributions there would be additional labels, for instancex2_label, etc. Furthermore, for plots there are the corresponding labels in LaTeX:x1_label_tex,y_label_texand so on. Finally,x1_unitcontains the physical units for each observable, typicallyGeV, andy_unitthe units of the (differential) cross section, typically inpb. - The value of the key
arxivgives us the corresponding arXiv identifier, in this case for the experimental measurement; enter the value into the search field of https://arxiv.org, and it’ll lead you to the paper. - The measured data itself you can get from the location specified by
hepdata, which is a digital object identifier (DOI). Go to https://www.doi.org/ and enter the string there. This will get you to the right table on https://www.hepdata.net for the corresponding observable. Together with the paper this will make clear that the predictions show the pseudorapidity of the positively-charged lepton, corresponding to table 6 in the paper. - The presence of
mg5amc_repoandmg5amc_revnoshow that Madgraph5_aMC@NLO was used to calculate the predictions. Since their values are empty, an official release was used, whose version we can read off fromruncard. If these values were non-zero, they’d point to the Madgraph5 repository and its revision number. - Finally, the value of
runcardcontains runcards of the calculation and also the information that Madgraph5_aMC@NLO v3.1.0 was used run the calculation, the values of all independent parameters and a few cuts.
If you’d like a complete description of all recognized metadata, have a look at the full list.
Orders, bins and channels
Each grid is—basically—a three-dimensional array of subgrids, which are the actual interpolation grids. The three dimensions are:
- orders (
o), - bins (
b) and - channels (
c).
You can use the subcommand read to see exactly how each grid is built. Let’s
go through them one by one using our grid:
pineappl read --orders LHCB_WP_7TEV.pineappl.lz4
The output is:
o order
-+----------------
0 O(a^2)
1 O(as^1 a^2)
2 O(as^1 a^2 lr^1)
3 O(as^1 a^2 lf^1)
4 O(a^3)
5 O(a^3 lr^1)
6 O(a^3 lf^1)
This shows that the grid contains:
- a leading order (LO) with index
0, which has the coupling alpha squared (a^2), - the QCD next-to-leading order (NLO) with index
1and - the EW NLO with index
4.
Additionally, there are
- NLO grids with factorization-log dependent terms, with indices
3and6. The corresponding - renormalization-logs (
2and5) are also present, but their contributions are in fact zero.
These last two subgrid types are needed if during the convolution the scale-variation uncertainty should be calculated.
Now let’s look at the bins:
pineappl read --bins LHCB_WP_7TEV.pineappl.lz4
which prints:
b etal norm
-+----+----+----
0 2 2.25 0.25
1 2.25 2.5 0.25
2 2.5 2.75 0.25
3 2.75 3 0.25
4 3 3.25 0.25
5 3.25 3.5 0.25
6 3.5 4 0.5
7 4 4.5 0.5
this shows the bin indices b for the observable etal, with their left and
right bin limits, which you’ve already seen in convolve. The column norm
shows the factor that all convolutions are divided with. Typically, as shown in
this case, this is the bin width, but in general this can be different.
Finally, let’s have a look at the channel definition:
pineappl read --channels LHCB_WP_7TEV.pineappl.lz4
This prints all partonic initial states that contribute to this process:
c entry entry
-+------------+------------
0 1 × ( 2, -1) 1 × ( 4, -3)
1 1 × (21, -3) 1 × (21, -1)
2 1 × (22, -3) 1 × (22, -1)
3 1 × ( 2, 21) 1 × ( 4, 21)
4 1 × ( 2, 22) 1 × ( 4, 22)
In this case you see that the up–anti-down (2, -1) and charm–anti-strange (4,
-3) initial states (the numbers are PDG MC IDs) are
grouped together in a single channel, each with a factor of 1. In general
this number can be different from 1, if the Monte Carlo decides to factor out
CKM values or electric charges, for instance, to group more contributions with
the same matrix elements together into a single channel. This is an
optimization step, as fewer channels result in a smaller grid file.
Note that channels with the transposed initial states, for instance anti-down—up, are merged with each other, which always works if the two initial-state hadrons are the same; this is an optimization step, also to keep the size of the grid files small.
All remaining channels are the ones with a gluon, 21, or with a photon, 22.
pineappl orders: What’s the size of each perturbative order?
In the previous section you saw that each order is stored separately, which means you can perform convolutions separately for each order:
pineappl orders LHCB_WP_7TEV.pineappl.lz4 CT18NNLO
which prints
b etal dsig/detal O(as^0 a^2) O(as^1 a^2) O(as^0 a^3)
[] [pb] [%] [%] [%]
-+----+----+-----------+-----------+-----------+-----------
0 2 2.25 7.7526895e2 100.00 17.63 -1.25
1 2.25 2.5 7.1092145e2 100.00 17.47 -1.13
2 2.5 2.75 6.1876958e2 100.00 18.07 -1.04
3 2.75 3 5.0017809e2 100.00 18.53 -0.92
4 3 3.25 3.7228440e2 100.00 19.12 -0.85
5 3.25 3.5 2.5236943e2 100.00 19.84 -0.81
6 3.5 4 1.1857770e2 100.00 21.32 -0.83
7 4 4.5 2.7740964e1 100.00 24.59 -1.01
By default all higher orders are shown relative to the sum of all LOs. However,
this can be changed using the switches --normalize or -n, which asks for
the orders you define as 100 per cent. If we’d like the numbers to be
normalized to NLO QCD, we’d run
pineappl orders --normalize=a2,a2as1 LHCB_WP_7TEV.pineappl.lz4 CT18NNLO
which will show
b etal dsig/detal O(as^0 a^2) O(as^1 a^2) O(as^0 a^3)
[] [pb] [%] [%] [%]
-+----+----+-----------+-----------+-----------+-----------
0 2 2.25 7.7526895e2 85.01 14.99 -1.06
1 2.25 2.5 7.1092145e2 85.13 14.87 -0.96
2 2.5 2.75 6.1876958e2 84.69 15.31 -0.88
3 2.75 3 5.0017809e2 84.37 15.63 -0.78
4 3 3.25 3.7228440e2 83.95 16.05 -0.71
5 3.25 3.5 2.5236943e2 83.45 16.55 -0.67
6 3.5 4 1.1857770e2 82.42 17.58 -0.69
7 4 4.5 2.7740964e1 80.26 19.74 -0.81
pineappl channels: What’s the size of each channel?
You can also show a convolution separately for each channel:
pineappl channels LHCB_WP_7TEV.pineappl.lz4 CT18NNLO
This will show the following table,
b etal c size c size c size c size c size
[] [%] [%] [%] [%] [%]
-+----+----+-+------+-+------+-+-----+-+----+-+----
0 2 2.25 0 111.00 3 -7.91 1 -3.10 2 0.00 4 0.00
1 2.25 2.5 0 111.83 3 -8.69 1 -3.15 2 0.00 4 0.00
2 2.5 2.75 0 112.68 3 -9.37 1 -3.31 2 0.00 4 0.00
3 2.75 3 0 113.49 3 -9.97 1 -3.53 2 0.00 4 0.00
4 3 3.25 0 114.26 3 -10.36 1 -3.90 2 0.00 4 0.00
5 3.25 3.5 0 115.00 3 -10.58 1 -4.42 2 0.00 4 0.00
6 3.5 4 0 115.65 3 -10.25 1 -5.39 2 0.00 4 0.00
7 4 4.5 0 115.81 3 -8.58 1 -7.23 2 0.00 4 0.00
The most important channel is 0, which is the up-type–anti-down-type
combination. The channels with gluons are much smaller and negative. Channels
with a photon are zero, because the PDF set that we’ve chosen doesn’t have a
photon PDF. Let’s try again with NNPDF31_nnlo_as_0118_luxqed (remember to
install the set first) as the PDF set:
b etal c size c size c size c size c size
[] [%] [%] [%] [%] [%]
-+----+----+-+------+-+------+-+-----+-+----+-+----
0 2 2.25 0 111.04 3 -7.84 1 -3.23 4 0.02 2 0.01
1 2.25 2.5 0 111.87 3 -8.62 1 -3.29 4 0.02 2 0.01
2 2.5 2.75 0 112.73 3 -9.31 1 -3.45 4 0.02 2 0.01
3 2.75 3 0 113.56 3 -9.90 1 -3.69 4 0.01 2 0.01
4 3 3.25 0 114.34 3 -10.30 1 -4.07 4 0.01 2 0.01
5 3.25 3.5 0 115.07 3 -10.52 1 -4.57 4 0.02 2 0.01
6 3.5 4 0 115.59 3 -10.18 1 -5.43 2 0.01 4 0.01
7 4 4.5 0 115.16 3 -8.23 1 -6.94 4 0.01 2 0.01
pineappl uncert: How large are the scale and PDF uncertainties?
Let’s calculate the scale and PDF uncertainties for our grid:
pineappl uncert --pdf --scale-env=7 LHCB_WP_7TEV.pineappl.lz4 NNPDF31_nnlo_as_0118_luxqed
This will show a table very similar to pineappl convolve:
b etal dsig/detal PDF central PDF 7pt-svar (env)
[] [pb] [%] [%]
-+----+----+-----------+-----------+-----+----+-------+-------
0 2 2.25 7.7651327e2 7.7650499e2 -1.01 1.01 -3.69 2.64
1 2.25 2.5 7.1011428e2 7.1008027e2 -1.01 1.01 -3.67 2.69
2 2.5 2.75 6.1683947e2 6.1679433e2 -1.01 1.01 -3.67 2.76
3 2.75 3 4.9791036e2 4.9786461e2 -1.03 1.03 -3.64 2.78
4 3 3.25 3.7016249e2 3.7012355e2 -1.07 1.07 -3.60 2.80
5 3.25 3.5 2.5055318e2 2.5052366e2 -1.13 1.13 -3.55 2.81
6 3.5 4 1.1746882e2 1.1745148e2 -1.33 1.33 -3.48 2.80
7 4 4.5 2.8023753e1 2.8018010e1 -4.05 4.05 -3.40 2.74
The first three columns are exactly the one that pineappl convolve shows. The
next columns are the PDF central predictions, and negative and positive PDF
uncertainties. These uncertainties are calculated using LHAPDF, so pineappl
always uses the correct algorithm no matter what type of PDF sets you use:
Hessian, Monte Carlo, etc. Note that we’ve chosen a PDF set with Monte Carlo
replicas which means that the results shown in dsig/detal and PDF central
aren’t exactly the same. The first number is calculated from the zeroth replica
of the set and the second number is the average of all replicas (except the
zeroth). You’ll notice that the PDF uncertainties are symmetric, which will not
necessarily be the case with Hessian PDF sets, for instance.
The two remaining columns show the scale-variation uncertainty which is
estimated from taking the envelope of a 7-point scale variation. By changing
the parameter of --scale-env to a different value (3 or 9), you can
change the scale-variation procedure.
pineappl pull: Are two PDF sets compatible with each other?
A variation of PDF uncertainties are pulls; they quantify how different predictions for two PDF sets are. The pull is defined as the difference of the two predictions $\sigma_1$ and $\sigma_2$, in terms of their PDF uncertainties $\delta \sigma_1$ and $\delta \sigma_2$:
\[\text{pull} = \frac{\sigma_2 - \sigma_1}{\sqrt{(\delta \sigma_1)^2 + (\delta \sigma_2)^2}}\]You can calculate it for each bin using:
pineappl pull LHCB_WP_7TEV.pineappl.lz4 CT18NNLO NNPDF31_nnlo_as_0118_luxqed
in which CT18NNLO will be used to calculate $\sigma_1$ and $\delta \sigma_1$
and NNPDF31_nnlo_as_0118_luxqed for $\sigma_2$ and $\delta \sigma_2$. This
will show not only the pull, in the column total, but also how this pull is
calculated using the different channels:
b etal total c pull c pull c pull c pull c pull
[] [σ] [σ] [σ] [σ] [σ] [σ]
-+----+----+------+-+------+-+------+-+------+-+-----+-+-----
0 2 2.25 0.065 0 0.086 1 -0.058 3 0.024 4 0.009 2 0.005
1 2.25 2.5 -0.037 1 -0.042 0 -0.029 3 0.024 4 0.006 2 0.004
2 2.5 2.75 -0.102 0 -0.097 1 -0.043 3 0.030 4 0.006 2 0.002
3 2.75 3 -0.149 0 -0.148 1 -0.045 3 0.036 4 0.005 2 0.004
4 3 3.25 -0.188 0 -0.190 1 -0.045 3 0.039 4 0.004 2 0.003
5 3.25 3.5 -0.235 0 -0.250 3 0.044 1 -0.037 4 0.005 2 0.002
6 3.5 4 -0.283 0 -0.344 3 0.050 1 0.004 2 0.003 4 0.003
7 4 4.5 0.144 0 0.073 3 0.037 1 0.031 4 0.002 2 0.001
Looking at the total column you can see that the numbers are much smaller
than 1, where 1 corresponds to a one sigma difference. This we expect
knowing that this dataset is used in the fit of both PDF sets. The remaining
columns show how different channels (with indices in the c column) contribute
to the total pull. For the last bin, for instance, we see channel 0
contributes roughly half to the total pull, the remaining pull coming from
channels 3 and 1.
Note that CT18NNLO doesn’t have a photon PDF, but the NNPDF set has one. However, for these observables the photon PDF contribution is too small to make a difference in the pull.
pineappl plot: Show me a plot of the predictions!
Often a good way to start understanding predictions is to plot them. Fortunately, this is easy with PineAPPL:
pineappl plot LHCB_WP_7TEV.pineappl.lz4 CT18NNLO > plot.py
This will write a matplotlib script in Python. Note that the script is
written to the standard output and redirected into plot.py. The advantage of
writing a plotting script instead of directly producing the plot is that you
can change it according to your needs. Finally, let’s run the plotting script:
python3 plot.py
This will create the file LHCB_WP_7TEV.pdf, which you can open. If you need a
different format than .pdf, look for the string '.pdf' in the plotting
script and change it to the file ending corresponding to your desired format.
Here’s how the result for .jpeg looks:
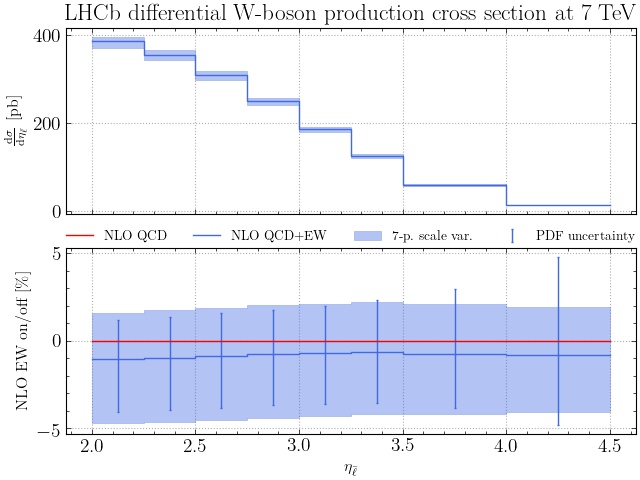
The plot subcommand is much more powerful, however. It accepts multiple PDF
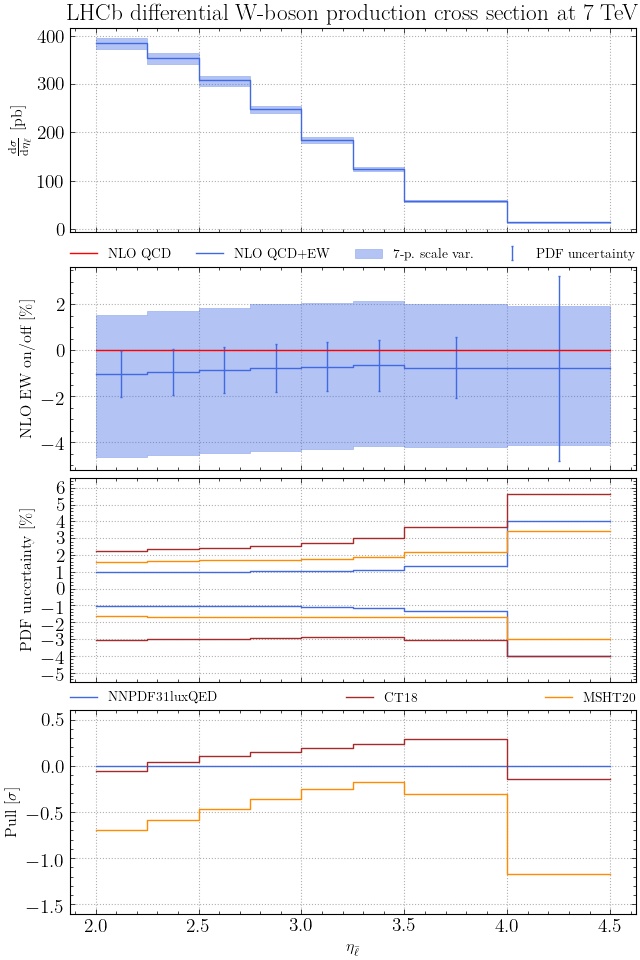
sets, for instance
pineappl plot LHCB_WP_7TEV.pineappl.lz4 NNPDF31_nnlo_as_0118_luxqed=NNPDF31luxQED \
CT18NNLO=CT18 MSHT20nnlo_as118=MSHT20 > plot.py
in which case more insets are plotted, which show the PDF uncertainty for
each set and also the pull using the first PDF set. As shown above, you can use
=plotlabel after the LHAPDF name to change the labels in the plot:
Note that you easily customize the generated script, plot.py in this case. It
is generated in such a way that style choices, plot labels and panel selection
is at the top of the script and separated from plotting routines and data,
which are found at the end of the file.
Conclusion
This is the end of the tutorial, but there are many more subcommands left that we didn’t discuss, and many more switches for subcommands that weren’t part of the tutorial. Remember that you can always run
pineappl
to get an overview of what’s there and what you might need, and that
pineappl <SUBCOMMAND> -h
gives you a more detailed description of the options and switches for each subcommand.